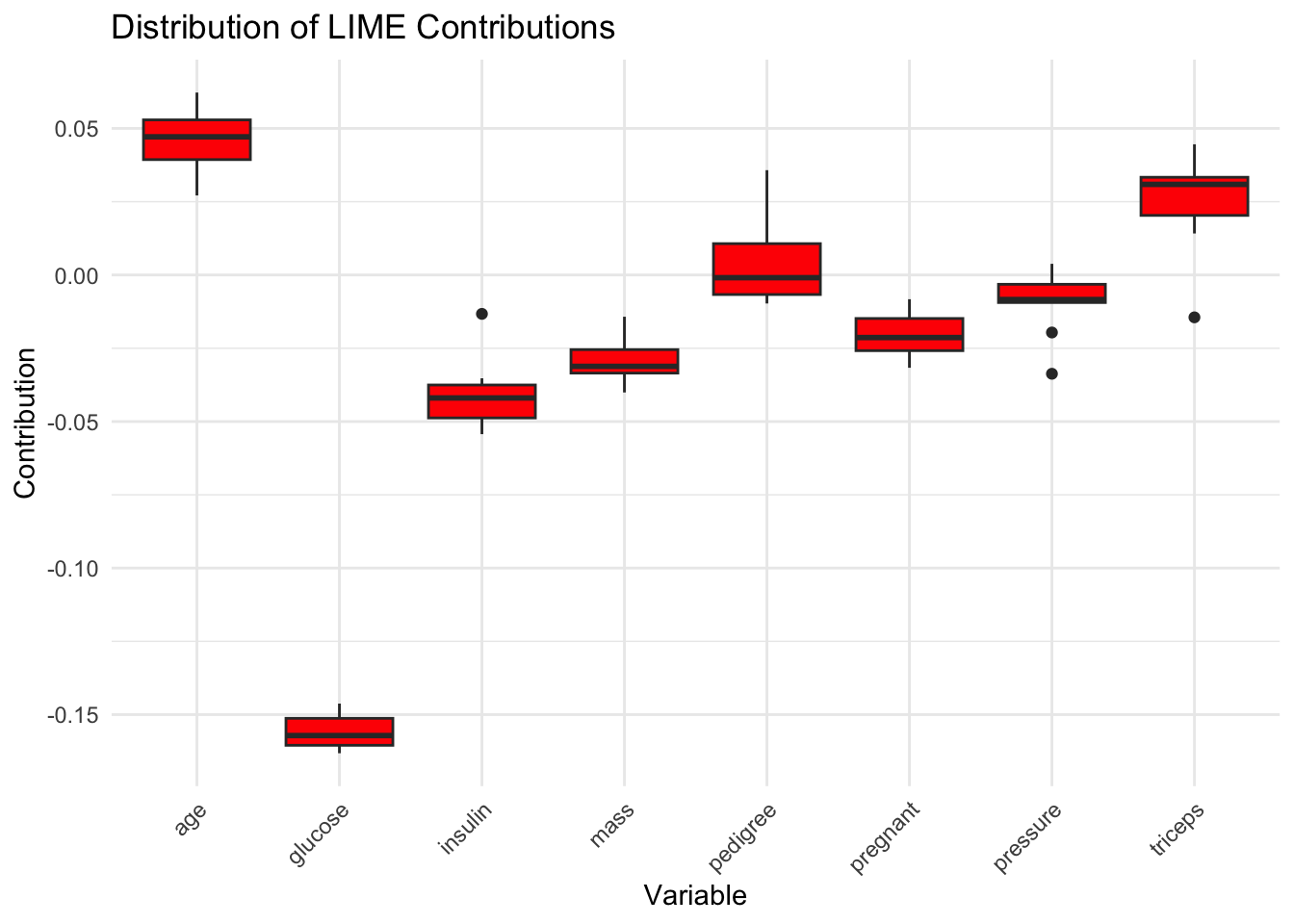
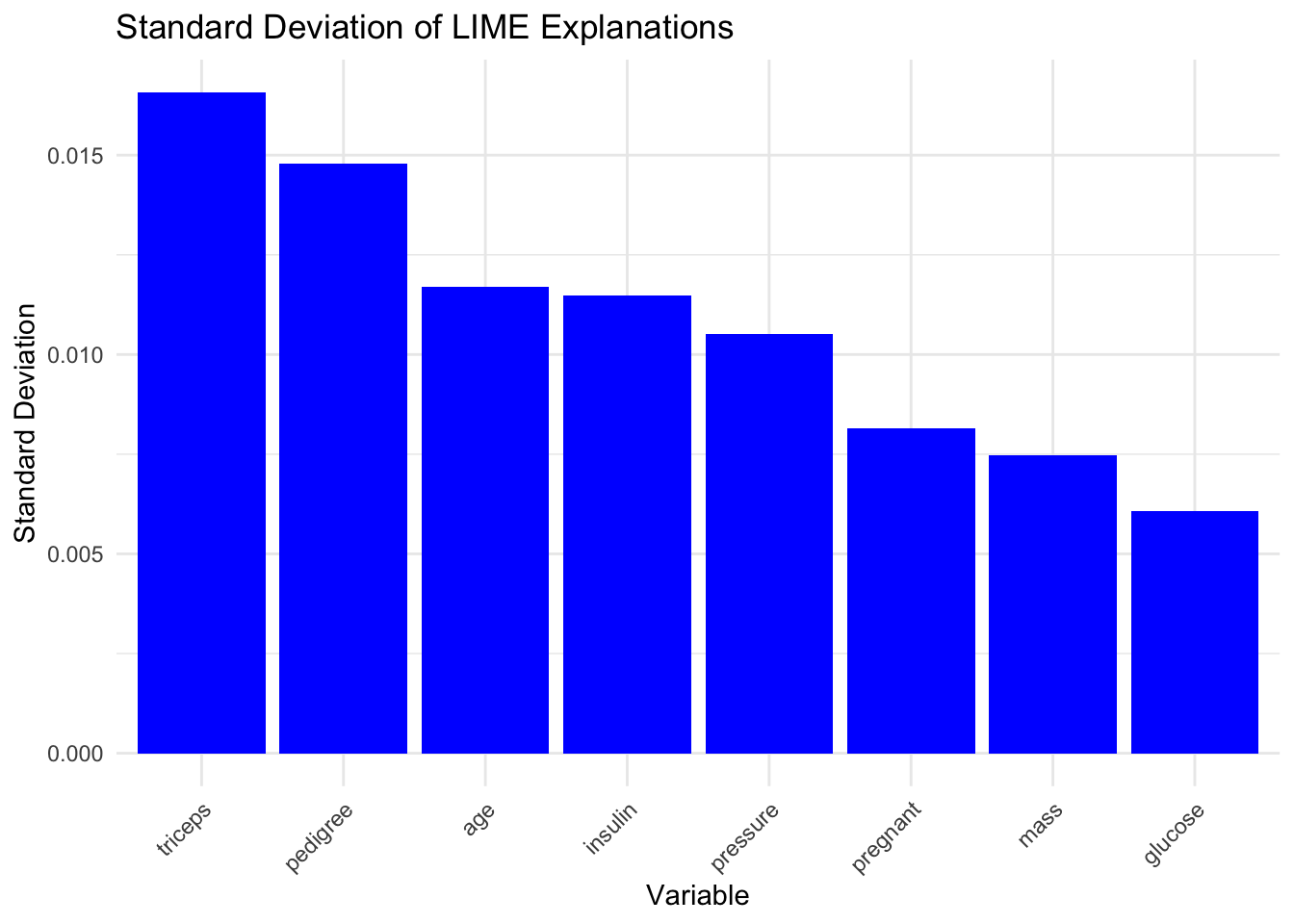
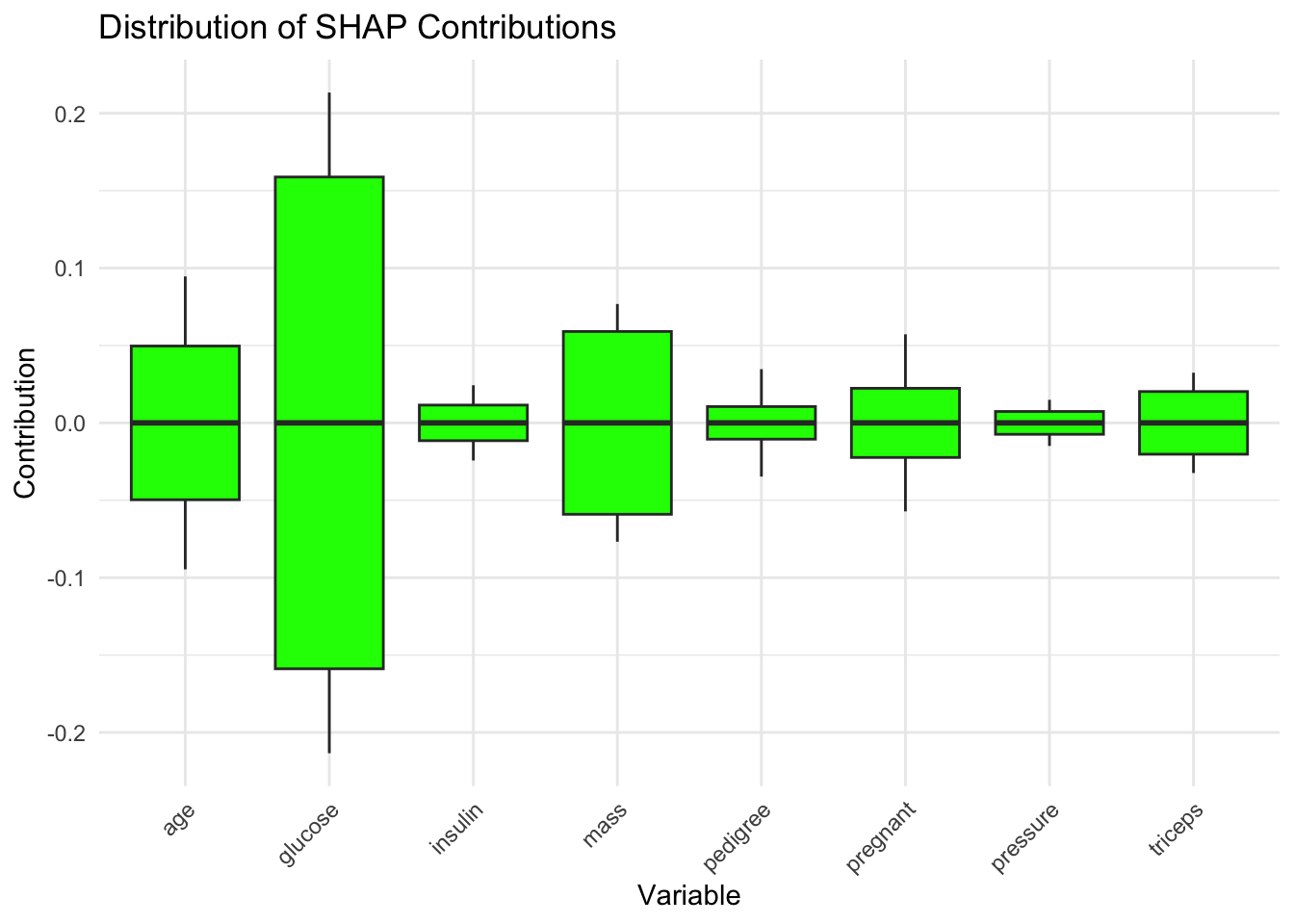
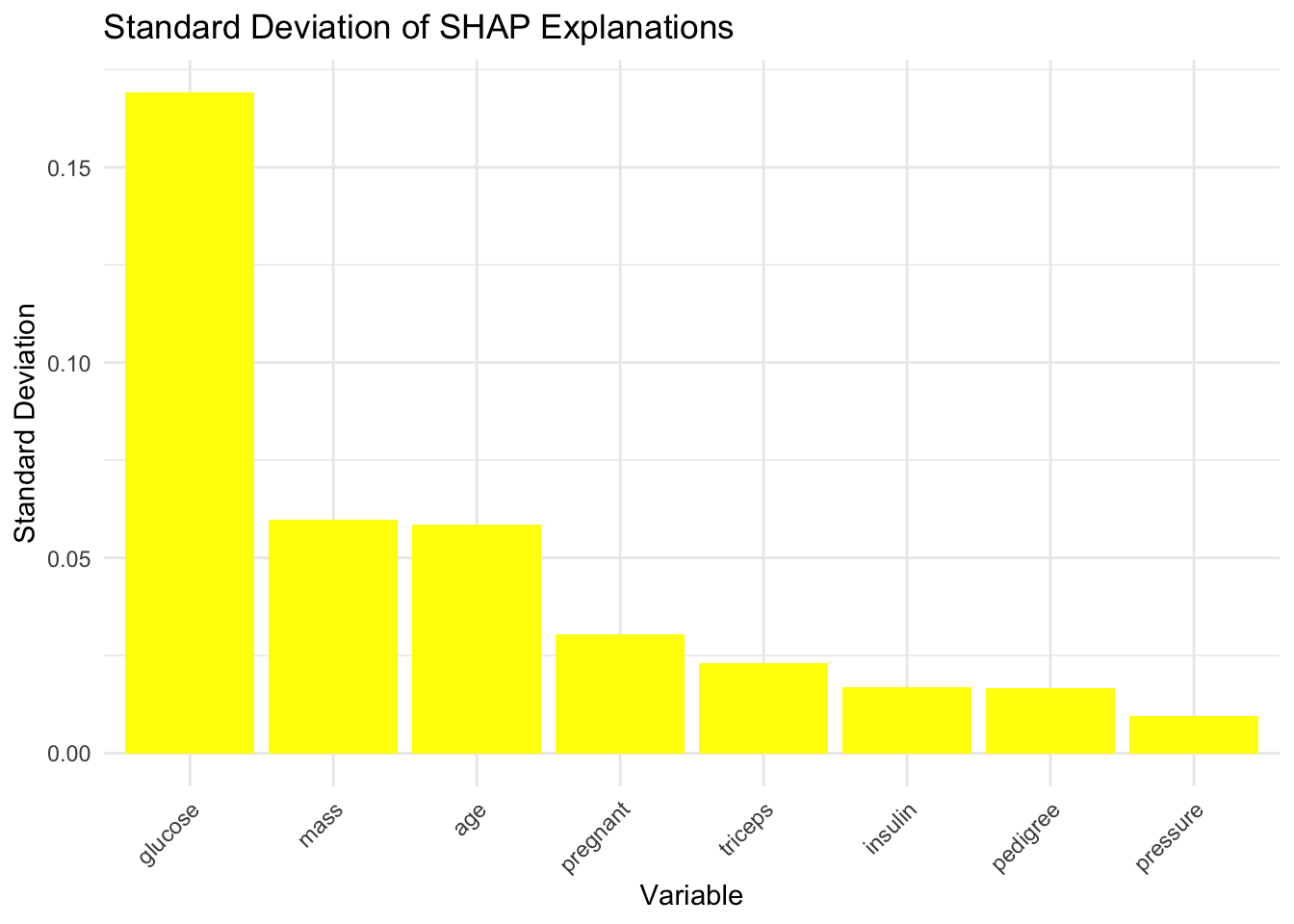
Code
library(randomForest)randomForest 4.7-1.2Type rfNews() to see new features/changes/bug fixes.Code
library(DALEX)Welcome to DALEX (version: 2.4.3).
Find examples and detailed introduction at: http://ema.drwhy.ai/Code
library(iml)
library(mlbench)
library(caret)Loading required package: ggplot2
Attaching package: 'ggplot2'The following object is masked from 'package:randomForest':
marginLoading required package: latticeCode
data(PimaIndiansDiabetes)
df <- na.omit(PimaIndiansDiabetes)
set.seed(5293)
train_idx <- createDataPartition(df$diabetes, p = 0.8, list = FALSE)
train_data <- df[train_idx, ]
test_data <- df[-train_idx, ]
lime_results <- list()
shap_results <- list()
for (i in 1:10) {
set.seed(i)
rf_model <- randomForest(diabetes ~ ., data = train_data, ntree = 100)
# LIME
explainer_lime <- DALEX::explain(
rf_model,
data = train_data[, -ncol(train_data)],
y = train_data$diabetes
)
lime_expl <- predict_parts(
explainer_lime,
new_observation = test_data[1, -ncol(test_data)],
type = "break_down"
)
lime_results[[i]] <- lime_expl
# SHAP
X <- train_data[, -ncol(train_data)]
predictor <- iml::Predictor$new(
rf_model,
data = X,
y = train_data$diabetes,
type = "prob"
)
shap <- iml::Shapley$new(predictor, x.interest = test_data[1, -ncol(test_data)])
shap_results[[i]] <- shap$results
}Preparation of a new explainer is initiated
-> model label : randomForest ( default )
-> data : 615 rows 8 cols
-> target variable : 615 values
-> predict function : yhat.randomForest will be used ( default )
-> predicted values : No value for predict function target column. ( default )
-> model_info : package randomForest , ver. 4.7.1.2 , task classification ( default )
-> model_info : Model info detected classification task but 'y' is a factor . ( WARNING )
-> model_info : By deafult classification tasks supports only numercical 'y' parameter.
-> model_info : Consider changing to numerical vector with 0 and 1 values.
-> model_info : Otherwise I will not be able to calculate residuals or loss function.
-> predicted values : numerical, min = 0 , mean = 0.3471545 , max = 0.99
-> residual function : difference between y and yhat ( default )Warning in Ops.factor(y, predict_function(model, data)): '-' not meaningful for
factors -> residuals : numerical, min = NA , mean = NA , max = NA
A new explainer has been created!
Preparation of a new explainer is initiated
-> model label : randomForest ( default )
-> data : 615 rows 8 cols
-> target variable : 615 values
-> predict function : yhat.randomForest will be used ( default )
-> predicted values : No value for predict function target column. ( default )
-> model_info : package randomForest , ver. 4.7.1.2 , task classification ( default )
-> model_info : Model info detected classification task but 'y' is a factor . ( WARNING )
-> model_info : By deafult classification tasks supports only numercical 'y' parameter.
-> model_info : Consider changing to numerical vector with 0 and 1 values.
-> model_info : Otherwise I will not be able to calculate residuals or loss function.
-> predicted values : numerical, min = 0 , mean = 0.3489431 , max = 0.98
-> residual function : difference between y and yhat ( default )Warning in Ops.factor(y, predict_function(model, data)): '-' not meaningful for
factors -> residuals : numerical, min = NA , mean = NA , max = NA
A new explainer has been created!
Preparation of a new explainer is initiated
-> model label : randomForest ( default )
-> data : 615 rows 8 cols
-> target variable : 615 values
-> predict function : yhat.randomForest will be used ( default )
-> predicted values : No value for predict function target column. ( default )
-> model_info : package randomForest , ver. 4.7.1.2 , task classification ( default )
-> model_info : Model info detected classification task but 'y' is a factor . ( WARNING )
-> model_info : By deafult classification tasks supports only numercical 'y' parameter.
-> model_info : Consider changing to numerical vector with 0 and 1 values.
-> model_info : Otherwise I will not be able to calculate residuals or loss function.
-> predicted values : numerical, min = 0 , mean = 0.3501789 , max = 0.99
-> residual function : difference between y and yhat ( default )Warning in Ops.factor(y, predict_function(model, data)): '-' not meaningful for
factors -> residuals : numerical, min = NA , mean = NA , max = NA
A new explainer has been created!
Preparation of a new explainer is initiated
-> model label : randomForest ( default )
-> data : 615 rows 8 cols
-> target variable : 615 values
-> predict function : yhat.randomForest will be used ( default )
-> predicted values : No value for predict function target column. ( default )
-> model_info : package randomForest , ver. 4.7.1.2 , task classification ( default )
-> model_info : Model info detected classification task but 'y' is a factor . ( WARNING )
-> model_info : By deafult classification tasks supports only numercical 'y' parameter.
-> model_info : Consider changing to numerical vector with 0 and 1 values.
-> model_info : Otherwise I will not be able to calculate residuals or loss function.
-> predicted values : numerical, min = 0 , mean = 0.3503252 , max = 0.98
-> residual function : difference between y and yhat ( default )Warning in Ops.factor(y, predict_function(model, data)): '-' not meaningful for
factors -> residuals : numerical, min = NA , mean = NA , max = NA
A new explainer has been created!
Preparation of a new explainer is initiated
-> model label : randomForest ( default )
-> data : 615 rows 8 cols
-> target variable : 615 values
-> predict function : yhat.randomForest will be used ( default )
-> predicted values : No value for predict function target column. ( default )
-> model_info : package randomForest , ver. 4.7.1.2 , task classification ( default )
-> model_info : Model info detected classification task but 'y' is a factor . ( WARNING )
-> model_info : By deafult classification tasks supports only numercical 'y' parameter.
-> model_info : Consider changing to numerical vector with 0 and 1 values.
-> model_info : Otherwise I will not be able to calculate residuals or loss function.
-> predicted values : numerical, min = 0 , mean = 0.3474309 , max = 0.99
-> residual function : difference between y and yhat ( default )Warning in Ops.factor(y, predict_function(model, data)): '-' not meaningful for
factors -> residuals : numerical, min = NA , mean = NA , max = NA
A new explainer has been created!
Preparation of a new explainer is initiated
-> model label : randomForest ( default )
-> data : 615 rows 8 cols
-> target variable : 615 values
-> predict function : yhat.randomForest will be used ( default )
-> predicted values : No value for predict function target column. ( default )
-> model_info : package randomForest , ver. 4.7.1.2 , task classification ( default )
-> model_info : Model info detected classification task but 'y' is a factor . ( WARNING )
-> model_info : By deafult classification tasks supports only numercical 'y' parameter.
-> model_info : Consider changing to numerical vector with 0 and 1 values.
-> model_info : Otherwise I will not be able to calculate residuals or loss function.
-> predicted values : numerical, min = 0 , mean = 0.3496748 , max = 0.99
-> residual function : difference between y and yhat ( default )Warning in Ops.factor(y, predict_function(model, data)): '-' not meaningful for
factors -> residuals : numerical, min = NA , mean = NA , max = NA
A new explainer has been created!
Preparation of a new explainer is initiated
-> model label : randomForest ( default )
-> data : 615 rows 8 cols
-> target variable : 615 values
-> predict function : yhat.randomForest will be used ( default )
-> predicted values : No value for predict function target column. ( default )
-> model_info : package randomForest , ver. 4.7.1.2 , task classification ( default )
-> model_info : Model info detected classification task but 'y' is a factor . ( WARNING )
-> model_info : By deafult classification tasks supports only numercical 'y' parameter.
-> model_info : Consider changing to numerical vector with 0 and 1 values.
-> model_info : Otherwise I will not be able to calculate residuals or loss function.
-> predicted values : numerical, min = 0 , mean = 0.3470244 , max = 0.98
-> residual function : difference between y and yhat ( default )Warning in Ops.factor(y, predict_function(model, data)): '-' not meaningful for
factors -> residuals : numerical, min = NA , mean = NA , max = NA
A new explainer has been created!
Preparation of a new explainer is initiated
-> model label : randomForest ( default )
-> data : 615 rows 8 cols
-> target variable : 615 values
-> predict function : yhat.randomForest will be used ( default )
-> predicted values : No value for predict function target column. ( default )
-> model_info : package randomForest , ver. 4.7.1.2 , task classification ( default )
-> model_info : Model info detected classification task but 'y' is a factor . ( WARNING )
-> model_info : By deafult classification tasks supports only numercical 'y' parameter.
-> model_info : Consider changing to numerical vector with 0 and 1 values.
-> model_info : Otherwise I will not be able to calculate residuals or loss function.
-> predicted values : numerical, min = 0 , mean = 0.3438699 , max = 0.98
-> residual function : difference between y and yhat ( default )Warning in Ops.factor(y, predict_function(model, data)): '-' not meaningful for
factors -> residuals : numerical, min = NA , mean = NA , max = NA
A new explainer has been created!
Preparation of a new explainer is initiated
-> model label : randomForest ( default )
-> data : 615 rows 8 cols
-> target variable : 615 values
-> predict function : yhat.randomForest will be used ( default )
-> predicted values : No value for predict function target column. ( default )
-> model_info : package randomForest , ver. 4.7.1.2 , task classification ( default )
-> model_info : Model info detected classification task but 'y' is a factor . ( WARNING )
-> model_info : By deafult classification tasks supports only numercical 'y' parameter.
-> model_info : Consider changing to numerical vector with 0 and 1 values.
-> model_info : Otherwise I will not be able to calculate residuals or loss function.
-> predicted values : numerical, min = 0 , mean = 0.3456748 , max = 0.99
-> residual function : difference between y and yhat ( default )Warning in Ops.factor(y, predict_function(model, data)): '-' not meaningful for
factors -> residuals : numerical, min = NA , mean = NA , max = NA
A new explainer has been created!
Preparation of a new explainer is initiated
-> model label : randomForest ( default )
-> data : 615 rows 8 cols
-> target variable : 615 values
-> predict function : yhat.randomForest will be used ( default )
-> predicted values : No value for predict function target column. ( default )
-> model_info : package randomForest , ver. 4.7.1.2 , task classification ( default )
-> model_info : Model info detected classification task but 'y' is a factor . ( WARNING )
-> model_info : By deafult classification tasks supports only numercical 'y' parameter.
-> model_info : Consider changing to numerical vector with 0 and 1 values.
-> model_info : Otherwise I will not be able to calculate residuals or loss function.
-> predicted values : numerical, min = 0 , mean = 0.3498374 , max = 0.99
-> residual function : difference between y and yhat ( default )Warning in Ops.factor(y, predict_function(model, data)): '-' not meaningful for
factors -> residuals : numerical, min = NA , mean = NA , max = NA
A new explainer has been created!