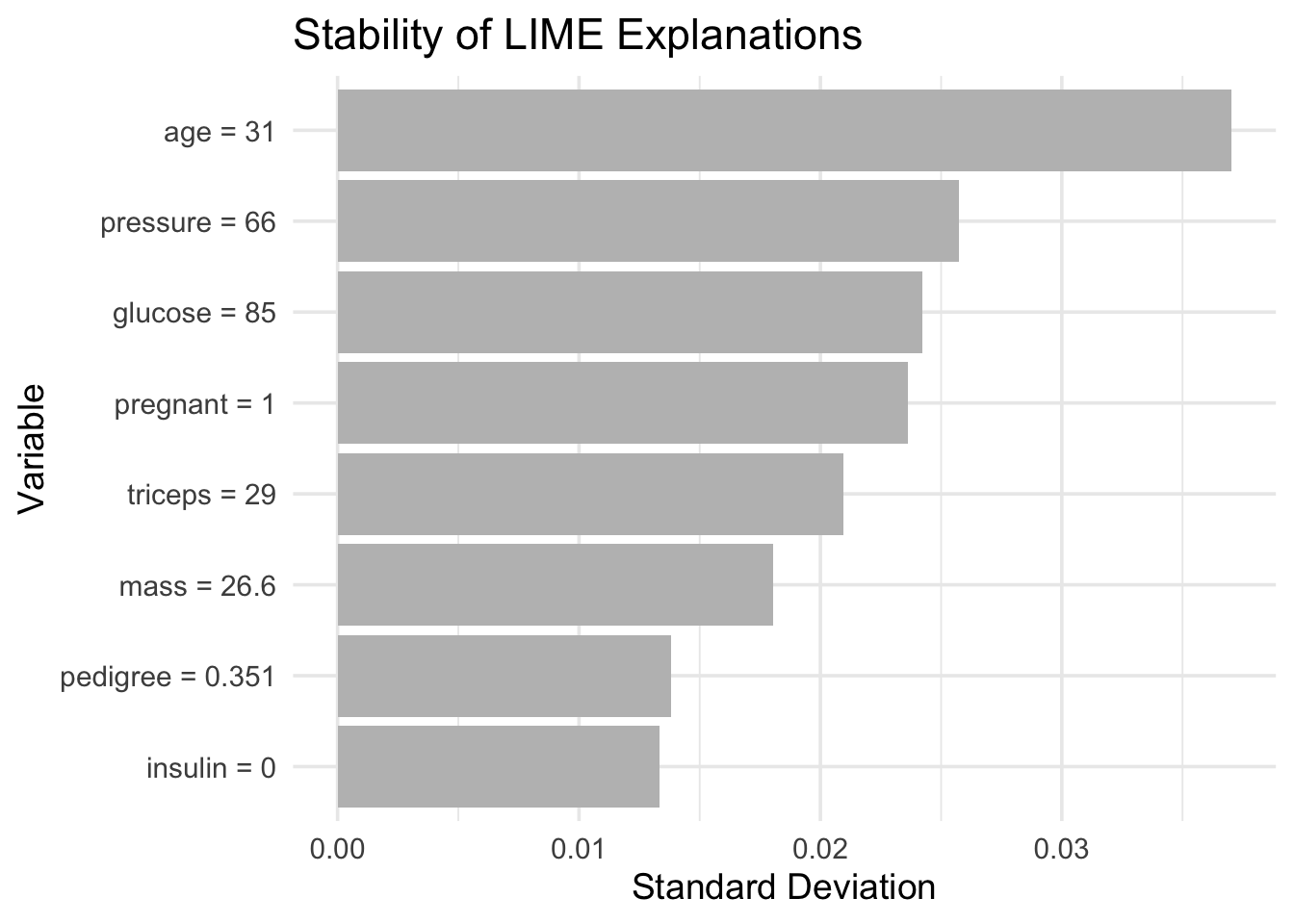
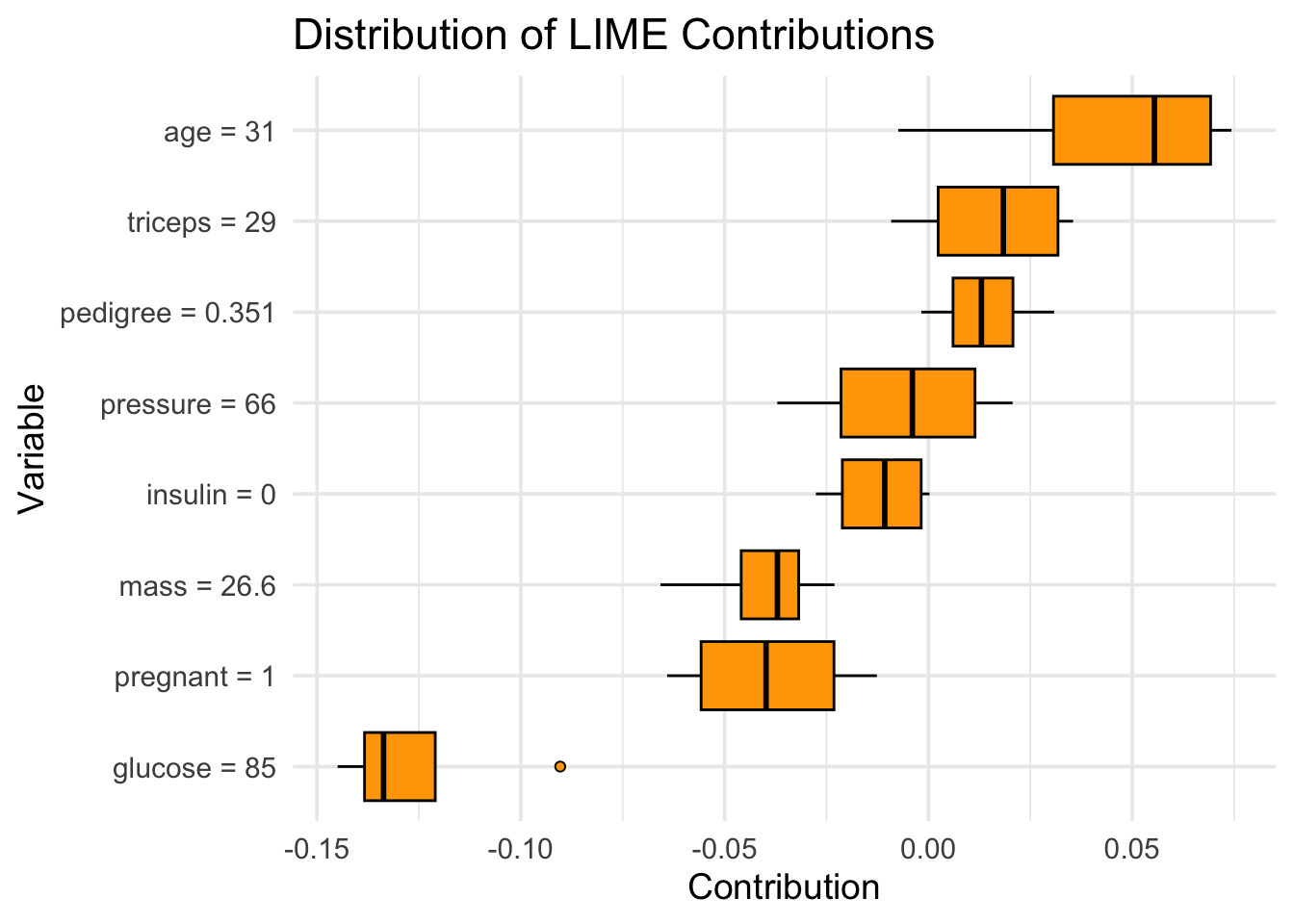
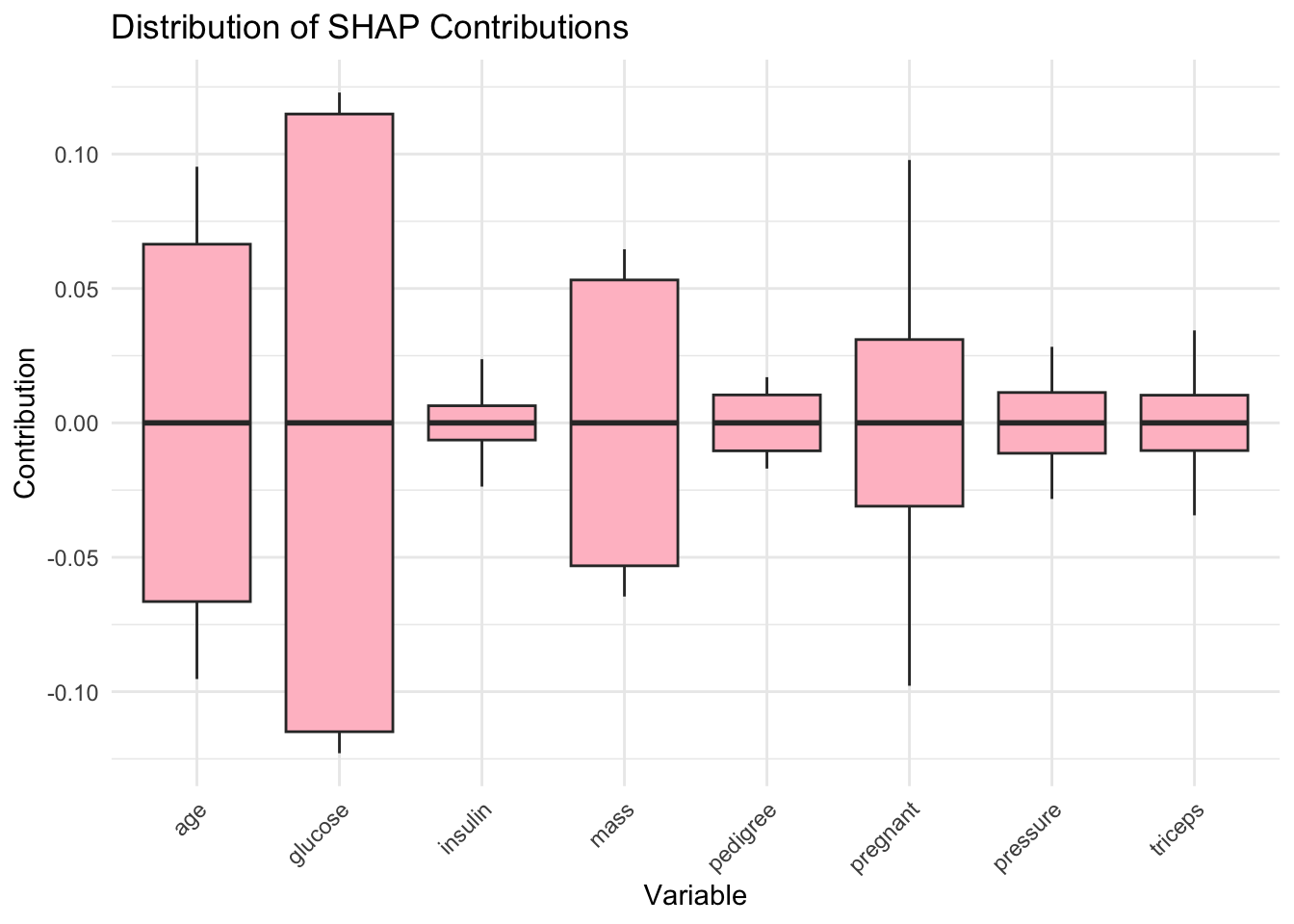
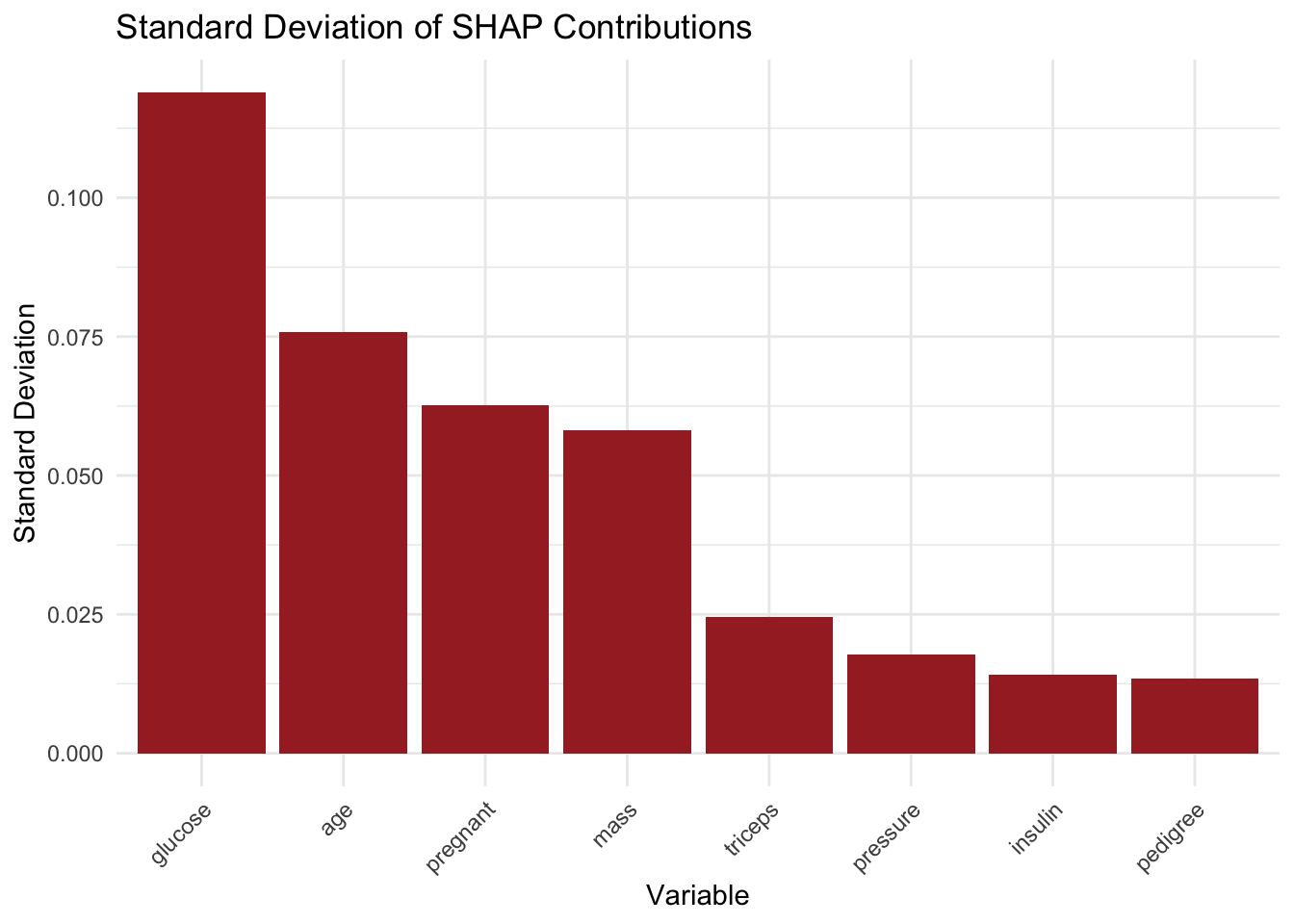
Preparation of a new explainer is initiated
-> model label : glm_size_0.1
-> data : 61 rows 8 cols
-> target variable : 61 values
-> predict function : yhat.glm will be used ( default )
-> predicted values : No value for predict function target column. ( default )
-> model_info : package stats , ver. 4.4.2 , task classification ( default )
-> predicted values : numerical, min = 0.01586211 , mean = 0.3442623 , max = 0.9887167
-> residual function : difference between y and yhat ( default )
-> residuals : numerical, min = -0.7908998 , mean = -1.719139e-11 , max = 0.8439318
A new explainer has been created!
Preparation of a new explainer is initiated
-> model label : glm_size_0.1
-> data : 61 rows 8 cols
-> target variable : 61 values
-> predict function : yhat.glm will be used ( default )
-> predicted values : No value for predict function target column. ( default )
-> model_info : package stats , ver. 4.4.2 , task classification ( default )
-> predicted values : numerical, min = 0.001370974 , mean = 0.4262295 , max = 0.9975288
-> residual function : difference between y and yhat ( default )
-> residuals : numerical, min = -0.7311193 , mean = 4.181597e-13 , max = 0.8912169
A new explainer has been created!
Preparation of a new explainer is initiated
-> model label : glm_size_0.1
-> data : 61 rows 8 cols
-> target variable : 61 values
-> predict function : yhat.glm will be used ( default )
-> predicted values : No value for predict function target column. ( default )
-> model_info : package stats , ver. 4.4.2 , task classification ( default )
-> predicted values : numerical, min = 0.001396533 , mean = 0.4262295 , max = 0.999766
-> residual function : difference between y and yhat ( default )
-> residuals : numerical, min = -0.8829128 , mean = 1.05795e-15 , max = 0.860971
A new explainer has been created!
Preparation of a new explainer is initiated
-> model label : glm_size_0.1
-> data : 61 rows 8 cols
-> target variable : 61 values
-> predict function : yhat.glm will be used ( default )
-> predicted values : No value for predict function target column. ( default )
-> model_info : package stats , ver. 4.4.2 , task classification ( default )
-> predicted values : numerical, min = 0.0002644932 , mean = 0.3278689 , max = 0.9819968
-> residual function : difference between y and yhat ( default )
-> residuals : numerical, min = -0.792514 , mean = -6.220705e-15 , max = 0.8393045
A new explainer has been created!
Preparation of a new explainer is initiated
-> model label : glm_size_0.1
-> data : 61 rows 8 cols
-> target variable : 61 values
-> predict function : yhat.glm will be used ( default )
-> predicted values : No value for predict function target column. ( default )
-> model_info : package stats , ver. 4.4.2 , task classification ( default )
-> predicted values : numerical, min = 0.007157103 , mean = 0.3442623 , max = 0.979401
-> residual function : difference between y and yhat ( default )
-> residuals : numerical, min = -0.7496494 , mean = 3.02266e-12 , max = 0.9041704
A new explainer has been created!
Preparation of a new explainer is initiated
-> model label : glm_size_0.3
-> data : 184 rows 8 cols
-> target variable : 184 values
-> predict function : yhat.glm will be used ( default )
-> predicted values : No value for predict function target column. ( default )
-> model_info : package stats , ver. 4.4.2 , task classification ( default )
-> predicted values : numerical, min = 0.0003898785 , mean = 0.3695652 , max = 0.9901829
-> residual function : difference between y and yhat ( default )
-> residuals : numerical, min = -0.9616683 , mean = -8.622124e-12 , max = 0.9118897
A new explainer has been created!
Preparation of a new explainer is initiated
-> model label : glm_size_0.3
-> data : 184 rows 8 cols
-> target variable : 184 values
-> predict function : yhat.glm will be used ( default )
-> predicted values : No value for predict function target column. ( default )
-> model_info : package stats , ver. 4.4.2 , task classification ( default )
-> predicted values : numerical, min = 0.009355782 , mean = 0.3315217 , max = 0.9668781
-> residual function : difference between y and yhat ( default )
-> residuals : numerical, min = -0.7974013 , mean = -5.942979e-14 , max = 0.9745445
A new explainer has been created!
Preparation of a new explainer is initiated
-> model label : glm_size_0.3
-> data : 184 rows 8 cols
-> target variable : 184 values
-> predict function : yhat.glm will be used ( default )
-> predicted values : No value for predict function target column. ( default )
-> model_info : package stats , ver. 4.4.2 , task classification ( default )
-> predicted values : numerical, min = 0.0009809545 , mean = 0.326087 , max = 0.9856423
-> residual function : difference between y and yhat ( default )
-> residuals : numerical, min = -0.8987656 , mean = -1.163467e-11 , max = 0.8936537
A new explainer has been created!
Preparation of a new explainer is initiated
-> model label : glm_size_0.3
-> data : 184 rows 8 cols
-> target variable : 184 values
-> predict function : yhat.glm will be used ( default )
-> predicted values : No value for predict function target column. ( default )
-> model_info : package stats , ver. 4.4.2 , task classification ( default )
-> predicted values : numerical, min = 0.0009854849 , mean = 0.3315217 , max = 0.9882061
-> residual function : difference between y and yhat ( default )
-> residuals : numerical, min = -0.9552123 , mean = -2.664226e-11 , max = 0.8874289
A new explainer has been created!
Preparation of a new explainer is initiated
-> model label : glm_size_0.3
-> data : 184 rows 8 cols
-> target variable : 184 values
-> predict function : yhat.glm will be used ( default )
-> predicted values : No value for predict function target column. ( default )
-> model_info : package stats , ver. 4.4.2 , task classification ( default )
-> predicted values : numerical, min = 0.009806256 , mean = 0.3315217 , max = 0.9836747
-> residual function : difference between y and yhat ( default )
-> residuals : numerical, min = -0.7999698 , mean = -2.986335e-14 , max = 0.9901937
A new explainer has been created!
Preparation of a new explainer is initiated
-> model label : glm_size_0.5
-> data : 307 rows 8 cols
-> target variable : 307 values
-> predict function : yhat.glm will be used ( default )
-> predicted values : No value for predict function target column. ( default )
-> model_info : package stats , ver. 4.4.2 , task classification ( default )
-> predicted values : numerical, min = 0.001861421 , mean = 0.3485342 , max = 0.9941528
-> residual function : difference between y and yhat ( default )
-> residuals : numerical, min = -0.9486772 , mean = -2.221301e-14 , max = 0.9856247
A new explainer has been created!
Preparation of a new explainer is initiated
-> model label : glm_size_0.5
-> data : 307 rows 8 cols
-> target variable : 307 values
-> predict function : yhat.glm will be used ( default )
-> predicted values : No value for predict function target column. ( default )
-> model_info : package stats , ver. 4.4.2 , task classification ( default )
-> predicted values : numerical, min = 0.003849884 , mean = 0.3192182 , max = 0.938876
-> residual function : difference between y and yhat ( default )
-> residuals : numerical, min = -0.8498261 , mean = -5.904496e-16 , max = 0.9838009
A new explainer has been created!
Preparation of a new explainer is initiated
-> model label : glm_size_0.5
-> data : 307 rows 8 cols
-> target variable : 307 values
-> predict function : yhat.glm will be used ( default )
-> predicted values : No value for predict function target column. ( default )
-> model_info : package stats , ver. 4.4.2 , task classification ( default )
-> predicted values : numerical, min = 0.000849326 , mean = 0.3420195 , max = 0.9861281
-> residual function : difference between y and yhat ( default )
-> residuals : numerical, min = -0.9336583 , mean = -2.586682e-13 , max = 0.9683198
A new explainer has been created!
Preparation of a new explainer is initiated
-> model label : glm_size_0.5
-> data : 307 rows 8 cols
-> target variable : 307 values
-> predict function : yhat.glm will be used ( default )
-> predicted values : No value for predict function target column. ( default )
-> model_info : package stats , ver. 4.4.2 , task classification ( default )
-> predicted values : numerical, min = 0.002316585 , mean = 0.3745928 , max = 0.9909388
-> residual function : difference between y and yhat ( default )
-> residuals : numerical, min = -0.9408385 , mean = -9.267045e-15 , max = 0.9686949
A new explainer has been created!
Preparation of a new explainer is initiated
-> model label : glm_size_0.5
-> data : 307 rows 8 cols
-> target variable : 307 values
-> predict function : yhat.glm will be used ( default )
-> predicted values : No value for predict function target column. ( default )
-> model_info : package stats , ver. 4.4.2 , task classification ( default )
-> predicted values : numerical, min = 0.001331084 , mean = 0.3289902 , max = 0.9995719
-> residual function : difference between y and yhat ( default )
-> residuals : numerical, min = -0.9344751 , mean = -5.301371e-11 , max = 0.995086
A new explainer has been created!
Preparation of a new explainer is initiated
-> model label : glm_size_1
-> data : 615 rows 8 cols
-> target variable : 615 values
-> predict function : yhat.glm will be used ( default )
-> predicted values : No value for predict function target column. ( default )
-> model_info : package stats , ver. 4.4.2 , task classification ( default )
-> predicted values : numerical, min = 0.001416797 , mean = 0.3495935 , max = 0.9943172
-> residual function : difference between y and yhat ( default )
-> residuals : numerical, min = -0.9518769 , mean = -4.522626e-14 , max = 0.989687
A new explainer has been created!
Preparation of a new explainer is initiated
-> model label : glm_size_1
-> data : 615 rows 8 cols
-> target variable : 615 values
-> predict function : yhat.glm will be used ( default )
-> predicted values : No value for predict function target column. ( default )
-> model_info : package stats , ver. 4.4.2 , task classification ( default )
-> predicted values : numerical, min = 0.001416797 , mean = 0.3495935 , max = 0.9943172
-> residual function : difference between y and yhat ( default )
-> residuals : numerical, min = -0.9518769 , mean = -4.522626e-14 , max = 0.989687
A new explainer has been created!
Preparation of a new explainer is initiated
-> model label : glm_size_1
-> data : 615 rows 8 cols
-> target variable : 615 values
-> predict function : yhat.glm will be used ( default )
-> predicted values : No value for predict function target column. ( default )
-> model_info : package stats , ver. 4.4.2 , task classification ( default )
-> predicted values : numerical, min = 0.001416797 , mean = 0.3495935 , max = 0.9943172
-> residual function : difference between y and yhat ( default )
-> residuals : numerical, min = -0.9518769 , mean = -4.522626e-14 , max = 0.989687
A new explainer has been created!
Preparation of a new explainer is initiated
-> model label : glm_size_1
-> data : 615 rows 8 cols
-> target variable : 615 values
-> predict function : yhat.glm will be used ( default )
-> predicted values : No value for predict function target column. ( default )
-> model_info : package stats , ver. 4.4.2 , task classification ( default )
-> predicted values : numerical, min = 0.001416797 , mean = 0.3495935 , max = 0.9943172
-> residual function : difference between y and yhat ( default )
-> residuals : numerical, min = -0.9518769 , mean = -4.522626e-14 , max = 0.989687
A new explainer has been created!
Preparation of a new explainer is initiated
-> model label : glm_size_1
-> data : 615 rows 8 cols
-> target variable : 615 values
-> predict function : yhat.glm will be used ( default )
-> predicted values : No value for predict function target column. ( default )
-> model_info : package stats , ver. 4.4.2 , task classification ( default )
-> predicted values : numerical, min = 0.001416797 , mean = 0.3495935 , max = 0.9943172
-> residual function : difference between y and yhat ( default )
-> residuals : numerical, min = -0.9518769 , mean = -4.522626e-14 , max = 0.989687
A new explainer has been created!